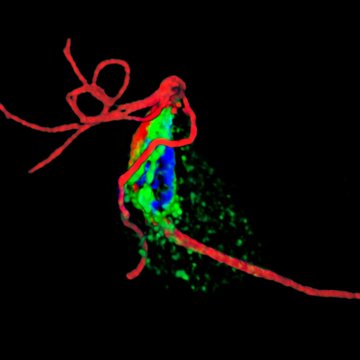
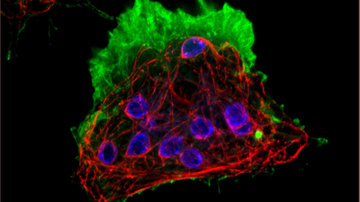
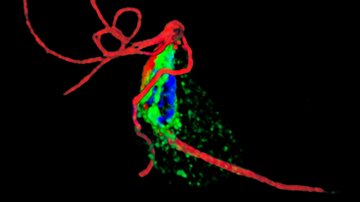
Verner Z., Žárský V., Le T., Narayanasamy R. K., Rada P., Rozbeský D., et al. (2021) Anaerobic peroxisomes in Entamoeba histolytica metabolize myo-inositol. PLoS Pathog 17(11): e1010041. https://doi.org/10.1371/journal.ppat.1010041
Tortorelli G., Pettollino F., Lai De-Hua, Tomčala A., Bacic A., Oborník M., Lukeš J., McFadden G. I. (2021). The cell wall polysaccharides of a photosynthetic relative of apicomplexans, Chromera velia. Journal of Phycology. https://doi.org/10.1111/jpy.13211
Petrů M., Dohnálek V., Füssy Z., Doležal P. Fates of sec, tat, and YidC translocases in mitochondria and other eukaryotic compartments. Molecular Biology and Evolution, Volume 38, Issue 12. December 2021. Pages 5241–5254. https://doi.org/10.1093/molbev/msab253
Blanvillain C., Saavedra S., Withers T., Votýpka J., Laroucau K., Lowenski S., Modrý D. (2021) Screening of diseases in wild exotic birds on Tahiti Island – implications for French Polynesian conservation. Pacific Conservation Biology 27, 284-295. https://doi.org/10.1071/PC20049
Tůmová P., Voleman L., Klingl A. et al. Inheritance of the reduced mitochondria of Giardia intestinalis is coupled to the flagellar maturation cycle. BMC Biol 19, 193 (2021). https://doi.org/10.1186/s12915-021-01129-7
Poláková E., Záhonová K., Albanaz A., Butenko A., Lukeš J., & Yurchenko V. (2021). Diverse telomeres in trypanosomatids. Parasitology, 148(10), 1254-1270. doi:10.1017/S0031182021000378
Kalčic F., Frydrych J., Doleželová E., Slapničková M., Pachl P., Poštová-Slavětínská L., Dračínský M., Hocková D., Zíková A., Janeba Z. (2021). C1´-Branched acyclic nucleoside phosphonates mimicking adenosine monophosphate: Potent inhibitors of Trypanosoma brucei adenine phosphoribosyltransferase. European Journal of Medicinal Chemistry: Volume 225, 113798 https://doi.org/10.1016/j.ejmech.2021.113798
Perner J., Hatalova T., Cabello-Donayre M., Urbanova V., Sojka D., Frantova H., Hartmann D., Jirsova D., Pérez-Victoria J. M. & Kopacek P. (2021). Haem-responsive gene transporter enables mobilization of host haem in ticks. Open Biology: Volume 11, Issue 9. https://doi.org/10.1098/rsob.210048
Kotál J., Polderdijk S. G. I., Langhansová H., Ederová M., Martins L. A., Beránková Z., Chlastáková A., Hajdušek O., Kotsyfakis M., Huntington J. A., Chmelař J. Ixodes ricinus salivary serpin Iripin-8 inhibits the intrinsic pathway of coagulation and complement. International Journal of Molecular Sciences. 2021; 22(17):9480. https://doi.org/10.3390/ijms22179480
Paris Z. (2021). Nuclear tRNA export in trypanosomes: A journey full of twists and turns guided by tRNA modifications. Parasitology, 148(10), 1219-1222. doi:10.1017/S0031182021000482
Kostygov A. Y., Grybchuk D., Kleschenko Y., Chistyakov D. S., Lukashev A. N., Gerasimov E. S., Yurchenko V. Analyses of Leishmania-LRV co-phylogenetic patterns and evolutionary variability of viral proteins. Viruses. 2021; 13(11):2305. https://doi.org/10.3390/v13112305
Poláková E., Albanaz A. T. S., Zakharova A., Novozhilova T. S., Gerasimov E. S., Yurchenko V. (2021) Ku80 is involved in telomere maintenance but dispensable for genomic stability in Leishmania mexicana. PLoS Negl Trop Dis 15(12): e0010041. https://doi.org/10.1371/journal.pntd.0010041
Zakharova A., Saura A., Butenko A., Podešvová L., Warmusová S., Kostygov A. Y., Nenarokova A., Lukeš J., Opperdoes F. R., Yurchenko V. (2021). A new model trypanosomatid, Novymonas esmeraldas: Genomic perception of its “Candidatus Pandoraea novymonadis“ endosymbiont. Eukaryotic Cells, https://doi.org/10.1128/mBio.01606-21
Vargová R., Wideman J. G., Derelle R., Klimeš V., Kahn R. A., Dacks J. B., Eliáš M. A eukaryote-wide perspective on the diversity and evolution of the ARF GTPase protein family. Genome Biology and Evolution, Volume 13, Issue 8, August 2021, evab157. https://doi.org/10.1093/gbe/evab157
Dixit S., Kessler A. C., Henderson J., Pan X., Zhao R., D'Almeida G. S., Kulkarni S., Rubio M. A. T., Hegedűsová E., Ross R. L., Limbach P. A., Green B. D., Paris Z., Alfonzo J.D. Dynamic queuosine changes in tRNA couple nutrient levels to codon choice in Trypanosoma brucei. Nucleic Acids Res. 2021 Dec 16;49(22):12986-12999. doi: 10.1093/nar/gkab1204. (IF=16.97)
Kopáček P., Šíma R., Perner J. (2021) An mRNA-based anti-tick vaccine catches ticks red-handed. Sci Transl Med. 2021 Nov 17;13(620):eabm2504. https://www.science.org/doi/10.1126/scitranslmed.abm2504
Šnebergerová P., Bartošová-Sojková P., Jalovecká M., Sojka D. (2021) Plasmepsin-like aspartyl proteases in Babesia. Pathogens Sep 26;10(10):1241. https://www.mdpi.com/2076-0817/10/10/1241
Stairs C. W., Táborský P., Salomaki E. D., Kolisko M., Pánek T., Eme L., Hradilová M., Vlček Č., Jerlström-Hultqvist J., Roger A. J., Čepička I. (2021) Anaeramoebae are a divergent lineage of eukaryotes that shed light on the transition from anaerobic mitochondria to hydrogenosomes. Curr Biol. 31:5605-5612.e5
Treitli S. C., Peña-Diaz P., Hałakuc P., Karnkowska A., Hampl V. High quality genome assembly of the amitochondriate eukaryote Monocercomonoides exilis. Microb Genom. 2021 Dec;7(12). doi: 10.1099/mgen.0.000745. PMID: 34951395.
Tonelli G. B., Andrade J. D., Campos A. M., Margonari C., Amaral A. R., Volf P., Shaw E. J., Hamilton J. G. C. (2021) Examination of the interior of sand fly (Diptera: Psychodidae) abdomen reveals novel cuticular structures involved in pheromone release: Discovering the manifold. PLOS NEGLECTED TROPICAL DISEASES 15, Article Number e0009733.
Bahia A. C., Barletta A. B. F., Pinto L. C., Orfanó A. S., Nacif-Pimenta R., Volfova V., Volf P., Secundino N. F. C., Fernandes F. F., Pimenta P. F. P. (2021) Morphological characterization of the antennal sensilla of the afrotropical sand fly, Phlebotomus duboscqi (Diptera: Psychodidae). Journal of Medical Entomology 58, 634–645.
Kykalova B., Ticha L., Volf P., Loza Telleria E. (2021) Phlebotomus papatasi antimicrobial peptides in larvae and females and a gut-specific defensin upregulated by Leishmania major infection. Microorganisms 9, 2307.
Sumova P., Sanjoba C., Willen L., Polanska N., Matsumoto Y., Noiri E., Paul S. K., Ozbel Y., Volf P. (2021) PpSP32-like protein as a marker of human exposure to Phlebotomus argentipes in Leishmania donovani foci in Bangladesh. International Journal for Parasitology 51: 1059-
Ticha L., Kykalova B., Sadlova J., Gramiccia M., Gradoni L., Volf P. (2021) Development of various Leishmania (Sauroleishmania) tarentolae strains in three Phlebotomus species. Microorganisms 9: 2256.
Husnik F., Tashyreva D., Boscaro V., Geoge E. E., Lukeš J., Keeling P. J. (2021). Bacterial and archaeal symbioses with protists. Current Biology: Volume 31, Issue 13, Pages R862-R877. https://doi.org/10.1016/j.cub.2021.05.049
Kulkarni S., Rubio M. A. T., Hegedüsová E., Ross R. L., Limbach P. A., Alfonzo J. D., Paris Z. Preferential import of queuosine-modified tRNAs into Trypanosoma brucei mitochondrion is critical for organellar protein synthesis. Nucleic Acids Research, Volume 49, Issue 14, 20 August 2021. Pages 8247–8260, https://doi.org/10.1093/nar/gkab567
Paris Z., Svobodová M., Kachale A., Horáková E., Nenarokova A. & Lukeš J. (2021). A mitochondrial cytidine deaminase is responsible for C to U editing of tRNATrp to decode the UGA codon in Trypanosoma brucei. RNA Biology, 18:sup1, 278-286. DOI: 10.1080/15476286.2021.1940445
Doleželová E., Klejch T., Špaček P. et al. Acyclic nucleoside phosphonates with adenine nucleobase inhibit Trypanosoma brucei adenine phosphoribosyltransferase in vitro. Sci Rep 11, 13317 (2021). https://doi.org/10.1038/s41598-021-91747-6
Smilansky V., Jirků M., Milner D. S., Ibáñez R., Gratwicke B., Nicholls A., Lukeš J., Chambouvet A. & Richards T. A. (2021). Expanded host and geographic range of tadpole associations with the Severe Perkinsea infection group. Volume 17, Issue 6. https://doi.org/10.1098/rsbl.2021.0166
Mishra A., Naseer Kaur J., McSkimming D. I., Hegedüsová E., Dubey A. P., Ciganda M., Paris Z., Read L. K. (2021). Selective nuclear export of mRNAs is promoted by DRBD18 in Trypanosoma brucei. https://doi.org/10.1111/mmi.14773
Richtová J., Sheiner L., Gruber A., Yang S. M., Kořený L., Striepen B., Oborník M. Using diatom and apicomplexan models to study the heme pathway of Chromera velia. International Journal of Molecular Sciences. 2021; 22(12):6495. https://doi.org/10.3390/ijms22126495
Cadena L. R., Gahura O., Panicucci B., Zíková A., Hashimi H. (2021). Mitochondrial contact site and cristae organization system and F1FO-ATP synthase crosstalk is a fundamental property of mitochondrial cristae. mSphere, e00327-21, Volume 6, Issue 3. https://journals.asm.org/doi/abs/10.1128/mSphere.00327-21
Sojka D., Šnebergerová P., Robbertse L. Protease inhibition—An established strategy to combat infectious diseases. International Journal of Molecular Sciences. 2021; 22(11):5762. https://doi.org/10.3390/ijms22115762
Záhonová K., Lax G., Sinha S. D. et al. Single-cell genomics unveils a canonical origin of the diverse mitochondrial genomes of euglenozoans. BMC Biol 19, 103 (2021). https://doi.org/10.1186/s12915-021-01035-y
Lopes A. L. K., Kriegová E., Lukeš J., Krieger M. A., Ludwig A. (2021) Distribution of Merlin in eukaryotes and first report of DNA transposons in kinetoplastid protists. PLoS ONE 16(5): e0251133. https://doi.org/10.1371/journal.pone.0251133
Oborník M. (2021) Enigmatic evolutionary history of porphobilinogen deaminase in eukaryotic phototrophs. Biology 10, no. 5: 386. https://doi.org/10.3390/biology10050386
Salomaki E. D., Terpis K. X., Rueckert S. et al. Gregarine single-cell transcriptomics reveals differential mitochondrial remodeling and adaptation in apicomplexans. BMC Biol 19, 77 (2021). https://doi.org/10.1186/s12915-021-01007-2
Pyrih J., Pánek T., Durante I. M., Rašková V., Cimrhanzlová K., Kriegová E., Tsaousis A. D., Eliáš M., Lukeš J. Vestiges of the bacterial signal recognition particle-based protein targeting in mitochondria. Molecular Biology and Evolution, Volume 38, Issue 8, August 2021, Pages 3170–3187, https://doi.org/10.1093/molbev/msab090
Bílý T., Sheikh S., Mallet A., Bastin P., Pérez-Morga D., Lukeš J., Hashimi H. (2021) Ultrastructural changes of the mitochondrion during the life cycle of Trypanosoma brucei. https://doi.org/10.1111/jeu.12846
Turnšek J., Brunson J. K., del Pilar Martinez Viedma M., Deerinck T. J., Horák A., Oborník M., Bielinski V. A., Allen A. E. (2021) Proximity proteomics in a marine diatom reveals a putative cell surface-to-chloroplast iron trafficking pathway. eLife 10:e52770. https://doi.org/10.7554/eLife.52770
Padilla-Mejia N. E., Koreny L., Holden J., Vancová M., Lukeš J., Zoltner M., Field M. C. A hub-and-spoke nuclear lamina architecture in trypanosomes. J Cell Sci. 2021 Jun 15;134(12):jcs251264.
Sharma P. P., Kumar S., Srivastava S., Srivastava M., Jee B., Gorobets N. Y., Kumar D., Kumar M., Asthana S., Zhang P., Poonam, Zoltner M., Rathi B. Computational study of novel inhibitory molecule, 1-(4-((2S,3S)-3-amino-2-hydroxy-4-phenylbutyl)piperazin-1-yl)-3-phenylurea, with high potential to competitively block ATP binding to the RNA dependent RNA polymerase of SARS-CoV-2 virus. J Biomol Struct Dyn. 2021 Jun 21:1-19.
Hammond M., Zoltner M., Garrigan J., Butterfield E., Varga V., Lukeš J., Field M. C. The distinctive flagellar proteome of Euglena gracilis illuminates the complexities of protistan flagella adaptation. New Phytol. 2021 Jul 22.
Vorel J., Cwiklinski K., Roudnický P., Ilgová J., Jedličková L., Dalton J. P., Mikeš L., Gelnar M., Kašný M. (2021): Eudiplozoon nipponicum (Monogenea, Diplozoidae) and its adaptation to haematophagy as revealed by transcriptome and secretome profiling. BMC Genomics 22: 274. DOI: 10.1186/s12864-021-07589-z [IF2020=3.969]
Soukal P., Hrdá Š., Karnkowska A., Milanowski R., Szabová J., Hradilová M., Strnad H., Vlček Č., Čepička I., Hampl V. Heterotrophic euglenid Rhabdomonas costata resembles its phototrophic relatives in many aspects of molecular and cell biology. Sci Rep. 2021 Jun 22;11(1):13070. doi: 10.1038/s41598-021-92174-3. PMID: 34158556; PMCID: PMC8219788.
Sulesco T., Kasap O. E., Halada P., Oguz G., Rusnac D., Gresova M., Alten B., Volf P., Dvorak V. (2021) Phlebotomine sand fly survey in the Republic of Moldova: species composition, distribution and host preferences. PARASITES & VECTORS 14: Article Number 371.
Corrales R. M., Vaselek S., Neish R., Berry L., Brunet C. D., Crobu L., Kuk N., Mateos-Langerak J., Robinson D. R., Volf P., Mottram J. C., Sterkers Y., Bastien P. (2021) The kinesin of the flagellum attachment zone in Leishmania is required for cell morphogenesis, cell division and virulence in the mammalian host. PLoS Pathogens 17: e1009666.
Telleria E. L., Tinoco-Nunes B., Lestinova T., De Avellar L. M., Tempone A. J., Pitaluga A. N., Volf P., Traub-Cseko Y. M. (2021) Lutzomyia longipalpis antimicrobial peptides: Differential expression during development and potential involvement in vector interaction with Microbiota and Leishmania. MICROORGANISMS 9: Article Number 1271.
Willen L., Basanez M. G., Dvorak V., Veriegh F. B. D., Aboagye F. T., Idun B., Osman M. E., Osei-Atweneboana M. Y., Courteney O., Volf P. (2021). Human immune response against salivary antigens of Simulium damnosus s.l.: A new epidemiological marker for exposure to blackfly bites in onchocerciasis endemic areas. PLoS Neglected Tropical Diseases 15: e0009512.
Kniha E., Dvorak V., Milchram M., Obwaller A. G., Kohsler M., Poeppl W., Antoniou M., Chaskopoulou A., Paronyan L., Stefanovski J., Mooseder G., Volf P., Walochnik J. (2021) Phlebotomus (Adlerius) simici NITZULESCU, 1931: first record in Austria and phylogenetic relationship with other Adlerius species. PARASITES & VECTORS 14: Article Number 20.
Kniha E., Milchram M., Dvorak V., Halada P., Obwaller A. G., Poeppl W., Mooseder G., Volf P., Walochnik J. (2021). Ecology, seasonality and host preferences of Austrian Phlebotomus (Transphlebotomus) mascittii Grassi, 1908, populations. PARASITES & VECTORS 14: Article Number 291.
Diniz J. A., Chaves M. M., Vaselek S., Magalhaes R. D. M., Ricci-Azevedo R., De Carvalho R. V. H., Lorenzon L. B., Ferreira T. R., Zamboni D., Walrad P. B., Volf P., Sacks D. L., Cruz A. K. (2021) Protein methyltransferase 7 deficiency in Leishmania major increases neutrophil associated pathology in murine model. PLOS NEGLECTED TROPICAL DISEASES 15: Article Number e0009230.
Baker N., Catta-Preta C. M. C., Neish R., Sadlova J., Powell B., Alves-Ferreira E. V. C., Geoghegan V., Carnielli J. B. T., Newling K., Hughes C., Vojtkova B., Anand J., Mihut A., Walrad P. B., Wilson L. G., Pitchford J. W., Volf P., Mottram J. C. (2021) Systematic functional analysis of Leishmania protein kinases identifies regulators of differentiation or survival. Nature Communications 12: 1244.
Schwabl P., Boite M. C., Bussotti G., Jacobs A., Andersson B., Moreira O., Freitas-Mesquita A. L., Meyer-Fernandes J. R., Telleria E. L., Traub-Cseko Y., Vaselek S., Lestinova T., Volf P., Morgado F. N., Porrozzi R., Llewellyn M., Spath G. F., Cupolillo E. (2021) Colonization and genetic diversification processes of Leishmania infantum in the Americas. COMMUNICATIONS BIOLOGY 4: Article Number 139.
Ashwin H., Sadlova J., Vojtkova B., Becvar T., Lypaczewski P., Schwartz E., Greensteed E., Van Boxlaer K., Parin M., Lipinski K. S., Parkash V., Matlashewski G., Layton A. M., Lacey C. J., Jaffe C. L., Volf P., Kay P. M. (2021). Characterization of a new Leishmania major strain for use in a controlled human infection model. Nature Communications 12: 215.
Sloan M. A., Sadlova J., Lestinova T., Sanders M. J., Cotton J. A., Volf P., Ligoxygakis P. (2021) The Phlebotomus papatasi systemic transcriptional response to trypanosomatid-contaminated blood does not differ from the non-infected blood meal. PARASITES & VECTORS 14: Article Number 15.
Becvar T., Vojtkova B., Siriyasatien P., Votypka J. Modry D., Jahn P., Bates P., Carpenter S., Volf P. & Sadlova J. (2021) Experimental transmission of Leishmania (Mundinia) parasites by biting midges (Diptera: Ceratopogonidae). PLoS Pathogens 17(6): e1009654.
Lesiczka P., Hrazdilová K., Majerová K., Fonville M., Sprong H., Hönig V., Hofmannová L., Papežík P., Růžek D., Zurek L., Votýpka J. & Modrý D. (2021). The role of peridomestic animals in the eco-epidemiology of Anaplasma phagocytophilum. Microbial Ecology (https://doi.org/10.1007/s00248-021-01704-z).
Majerová K., Gutiérrez R., Fonville K., Hönig V., Papežík P., Hofmannová L., Lesiczka P. M., Nachum-Biala Y., Růžek D., Sprong H., Harrus S., Modrý D. & Votýpka J. (2021) Hedgehogs and squirrels as hosts of zoonotic Bartonella species. Pathogens 10: 686.
Votýpka J., Petrželková K. J., Brzoňová J., Jirků M., Modrý D. & Lukeš J. (2021) How monoxenous trypanosomatids revealed hidden feeding habits of their tsetse fly hosts. Folia Parasitologica 68: 019.
Belinky F., Ganguly I., Poliakov E., Yurchenko V., Rogozin I. B. Analysis of stop codons within prokaryotic protein-coding genes suggests frequent readthrough events. Int. J. Mol. Sci., 2021, 22: 1876.
Lukeš J., Tesařová M., Yurchenko V., Votýpka J. Characterization of a new cosmopolitan genus of trypanosomatid parasites, Obscuromonas gen. nov. (Blastocrithidiinae subfam. nov.). Eur. J. Protistol., 2021, 79: 125778.
Sádlová J., Podešvová L., Bečvář T., Bianchi C., Gerasimov E. S., Saura A., Glanzová K., Leštinová T., Matveeva N. S., Chmelová L., Mlacovská D., Spitzová T., Vojtková B., Volf P., Yurchenko V., Kraeva N. Catalase impairs Leishmania mexicana development and virulence. Virulence, 2021, 12(1): 852-867.
Charyyeva A., Çetinkaya Ü., Özkan B., Şahin S., Yaprak N., Şahin I., Yurchenko V., Kostygov A. Y. Genetic diversity of Leishmania tropica: unexpectedly complex distribution pattern. Acta Trop., 2021, 218: 105888.
Albanaz A. T. S., Gerasimov E. S., Shaw J. J., Sádlová J., Lukeš J., Volf P., Opperdoes F. R., Kostygov A. Y., Butenko A., Yurchenko V. Genome analysis of Endotrypanum and Porcisia spp., closest phylogenetic relatives of Leishmania, highlights the role of amastins in shaping pathogenicity. Genes. 2021, 12(3): 444.
Gerasimov E. S., Gasparyan A. A., Afonin D. A., Zimmer S. L., Kraeva N., Lukeš J., Yurchenko V., Kolesnikov A. Complete minicircle genome of Leptomonas pyrrhocoris reveals a source of its non-canonical mitochondrial RNA editing events. Nucl. Acids Res., 2021, 49(6): 3354-3370.
Frolov A. O., Kostygov A. Y., Yurchenko V. Development of monoxenous trypanosomatids and phytomonads in insects. Trends Parasitol., 2021, 37(6): 538-551.
Skalický T., Alves J. M. P., Morais A. C., Režnarová J., Butenko A., Lukeš J., Serrano M. G., Buck G. A., Teixeira M. M. G., Camargo E. P., Sanders M., Cotton J. A., Yurchenko V., Kostygov A. Y. Endosymbiont capture, a repeated process of endosymbiont transfer with replacement in trypanosomatids Angomonas spp. Pathogens, 2021, 10: 702.
Konorov E. A., Yurchenko V., Patraman I. V., Lukashev A. N., Oyun N. Y. The effects of genetic drift and genomic selection on differentiation and local adaptation of the introduced populations of Aedes albopictus in southern Russia. Peer J., 2021, 9: e11776.
Barcytė D., Eikrem W., Engesmo A., Seoane S., Wohlmann J., Horák A., Yurchenko T., Eliáš M. Olisthodiscus represents a new class of Ochrophyta. J. Phycol., 2021, 57: 1094-1118.
Tice A. K., Žihala D., Pánek T., Jones R. J., Salomaki E., Nenarokov S., Burki F., Eliáš M., Eme L., Roger A. J., Rokas A., Shen X.-X., Strassert J. F. H., Kolísko M., Brown M. W. PhyloFisher: a phylogenomic package for resolving deep eukaryotic relationships. PLoS Biol., 2021, 19: e3001365.
Larue G. E., Eliáš M., Roy S. W. Expansion and transformation of the minor spliceosomal system in the slime mold Physarum polycephalum. Curr. Biol., 2021, 31: P3125-3131.E4.
Herman E. K., Greninger A., van der Giezen M., Ginger M. L., Ramirez-Macias I., Miller H. C., Morgan M. J., Tsaousis A. D., Velle K., Vargová R., Záhonová K., Najle S. R., MacIntyre G., Muller N., Wittwer M., Zysset-Burri D. C., Eliáš M., Slamovits C., Weirauch M., Fritz-Laylin L., Marciano-Cabral F., Puzon G. J., Walsh T., Chiu C., Dacks J. B. Genomics and transcriptomics yields a systems-level view of the biology of the pathogen Naegleria fowleri. BMC Biol., 2021, 19: 142.
Pyrih J., Žárský V., Fellows D.J., Grosche C., Wloga D., Striepen B, Maier G.U., Tachezy J. 2021. The iron-sulfur scaffold protein HCF101 unveils the complexity of organellar evolution in SAR, Haptista and Cryptista. BMC Evolutionary Biology 21, Article number: 46 (2021)
Žárský V., Klimeš V., Pačes J., Vlček Č, Hradilová M., Beneš V., Nývltová E., Hrdý I., Pyrih J, Mach J., Barlow L., Stairs C.W., Eme L., Hall N., Eliáš M., Dacks BJ, Roger A., Tachezy J. 2021. The Mastigamoeba balamuthi genome and the nature of the free-living ancestor of Entamoeba. Mol. Biol. Evol. doi: 10.1093/molbev/msab020. Online ahead of print. PMID: 33528570, IF 11,062
Bensaoud Ch., Martins L. A., Aounallah H., Hackenberg M., Kotsyfakis M. Emerging roles of non-coding RNAs in vector-borne infections. J Cell Sci (2021) 134 (5): jcs246744. https://doi.org/10.1242/jcs.246744
Villafraz O., Biran M., Pineda E., Plazolles N., Cahoreau E., Ornitz Oliveira Souza R., et al. (2021) Procyclic trypanosomes recycle glucose catabolites and TCA cycle intermediates to stimulate growth in the presence of physiological amounts of proline. PLoS Pathog 17(3): e1009204. https://doi.org/10.1371/journal.ppat.1009204
Kostygov A.Y., Karnkowska A., Votýpka J., Tashyreva D., Maciszewski K., Yurchenko V. & Lukeš J. (2021) Euglenozoa: taxonomy, diversity and ecology, symbioses and viruses. Open Biology 11: 200407.
Horváthová L., Žárský V., Pánek T., Derelle R., Pyrih J., Motyčková A., Klápšťová V., Vinopalová M., Marková L., Voleman L., Klimeš V., Petrů M., Vaitová Z., Čepička I., Hryzáková K., Harant K., Gray M. W., Chami M., Guilvout I., Francetic O., Franz Lang B., Vlček Č., Tsaousis A.D., Eliáš M., Doležal P. Analysis of diverse eukaryotes suggests the existence of an ancestral mitochondrial apparatus derived from the bacterial type II secretion system. Nat Commun. 2021 May 19;12(1):2947. doi: 10.1038/s41467-021-23046-7.
Rout S., Oeljeklaus S., Makki A., Tachezy J., Warscheid B., Schneider A. Determinism and contingencies shaped the evolution of mitochondrial protein import. Proceedings of the National Academy of Sciences Feb 2021, 118 (6) e2017774118; DOI: 10.1073/pnas.2017774118
Hierro-Yap C., Šubrtová K., Gahura O., Panicucci B., Dewar C., Chinopoulos Ch., Schnaufer A., Zíková A. Bioenergetic consequences of FoF1–ATP synthase/ATPase deficiency in two life cycle stages of Trypanosoma brucei, Journal of Biological Chemistry, Volume 296, 2021 , ISSN 0021-9258, https://doi.org/10.1016/j.jbc.2021.100357.
Procházková M., Füzik T., Grybchuk D., Luca Falginell F., Podešvová L., Yurchenko V., Vácha R., Plevka P. Capsid structure of Leishmania RNA Virus 1. Journal of Virology. Jan 2021, 95 (3) e01957-20, doi:10.1128/JVI.01957-20
Čermáková, P., Maďarová, A., Baráth, P., Bellová, J., Yurchenko, V., & Horváth, A. (2021). Differences in mitochondrial NADH dehydrogenase activities in trypanosomatids. Parasitology, 1-10. doi:10.1017/S0031182020002425
Gahura O., Hierro Yap C., Zíková A. (2021). Redesigned and reversed: architectural and functional oddities of the trypanosomal ATP synthase. Parasitology. 8:1-41
Tobiasson V., Gahura O., Aibara S., Baradaran R., Zíková A., Amunts A. (2021). Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit. EMBO J.
Opperdoes F. R., Butenko A., Zakharova A., Gerasimov E. S., Zimmer S. L., Lukeš J., & Yurchenko V. (2021). The remarkable metabolism of Vickermania ingenoplastis: Genomic predictions. Pathogens, 10(1), 68. https://doi.org/10.3390/pathogens10010068
Butenko A., Hammond M., Field M. C., Ginger M. L., Yurchenko V., & Lukeš J. (2021). Reductionist pathways for parasitism in Euglenozoans? Expanded datasets provide new insights. Trends in parasitology, 37(2), 100–116. https://doi.org/10.1016/j.pt.2020.10.001
Mahmood S., Sima R., Urbanova V., Trentelman J. J. A., Krezdorn N., Winter P., Kopacek P., Hovius J. W., Hajdusek O. (2021). Identification of tick Ixodes ricinus midgut genes differentially expressed during the transmission of Borrelia afzelii spirochetes using a transcriptomic approach. Front Immunol. 11:612412. doi: 10.3389/fimmu.2020.612412.
Kozelková T., Doležel D., Grunclová L., Kučera M., Perner J., Kopáček P. (2021) Functional characterization of the insulin signaling pathway in the hard tick Ixodes ricinus. Ticks and Tick-borne Diseases 12 101694, https://doi.org/10.1016/j.ttbdis.2021.101694
Klouwens M. J., Trentelman J. J., Ersoz J. I., Nieves Marques Porto F., Sima R., Hajdusek O., Thakur M., Pal U., Hovius J. W. (2021) Investigating BB0405 as a novel Borrelia afzelii vaccination candidate in Lyme borreliosis. Sci Rep. 11(1):4775. doi: 10.1038/s41598-021-84130-y.
Dorrah M., Bensaoud C., Mohamed A. A., Sojka D., Bassal T. T. M., Kotsyfakis M. (2021) Comparison of the hemolysis machinery in two evolutionarily distant blood-feeding arthropod vectors of human diseases. PLoS Negl Trop Dis 15(2): e0009151. https://doi.org/10.1371/journal.pntd.0009151
Ahmad P., Bensaoud C., Mekki I., Rehman M. U., Kotsyfakis M. (2021). Long non-coding RNAs and their potential roles in the vector-host-pathogen triad. Life (Basel) 11(1):56. doi: 10.3390/life11010056.
Kotabova E., Malych R., Pierella Karlusich J. J., Kazamia E., Eichner M., Mach J., Lesuisse E., Bowler C., Prášil O., Sutak R. Complex response of the chlorarachniophyte Bigelowiella natans to iron availability. mSystems. 2021
Füssy Z., Vinopalová M., Treitli S. C., Pánek T., Smejkalová P., Čepička I., Doležal P., Hampl V. Retortamonads from vertebrate hosts share features of anaerobic metabolism and pre-adaptations to parasitism with diplomonads. Parasitol Int. 2021 Feb 21;82:102308. doi: 10.1016/j.parint.2021.102308.