About us
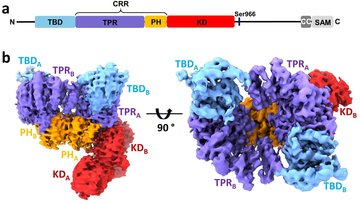
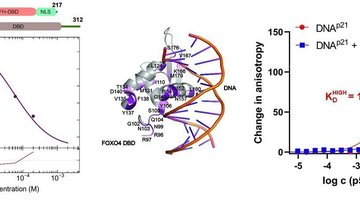
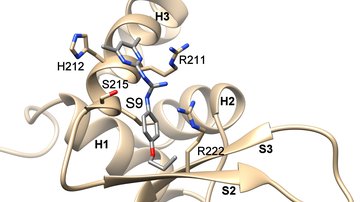
Our group focuses on studying protein-protein interactions with the aim to elucidate the molecular basis of regulatory mechanisms of signaling proteins and their complexes from the biologically important signaling pathways. The main focus is on 14-3-3 protein complexes with proteins involved in the regulation of apoptosis, cancer, G-protein and calcium signaling pathways.
We employ various biophysical (fluorescence spectroscopy, analytical ultracentrifugation, ITC, MST, DSF), structural (X-ray crystallography, NMR, SAXS, HDX-MS) and biochemical (site-directed mutagenesis, enzyme kinetics) approaches to understand the structure-function relationships of studied proteins and protein-protein complexes.
Objectives
- Preparation, biochemical and biophysical characterization of selected signaling proteins.
- Determination of binding affinities and stoichiometries of studied signaling complexes.
- Structural studies of selected signaling protein complexes.
Group Profile (Home Institution): Structural Biology of Signaling Proteins - Institute of Physiology AS CR, v.v.i. (cas.cz)