About us
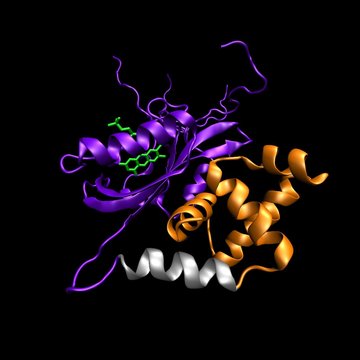
The aim of this Project is to understand how light controls the structure, dynamics and activity of photosensitive proteins.
Mission and vision
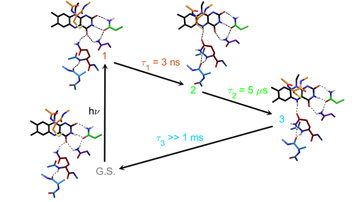
The aim of this Project is to understand how light controls the structure, dynamics and activity of photosensitive proteins. We focus on transcription factors that interact with specific sequences of DNA in a light-dependent manner (EL222, CarH, CooA). Photon absorption by the embedded chromophores triggers conformational changes in the protein chain that ultimately alter protein-protein and protein-DNA interaction networks in the cell. Our plan is to unveil the molecular details of photoswitching between “dark” and “light” states on time scales ranging from femtoseconds to hours. The acquired knowledge will allow us to create novel photoreceptors for use in areas like synthetic biology, photopharmacology and optogenetics.
Methods
We employ an integrative 4D structural biology approach combining methods with distinct spatial and temporal resolution:
- Molecular biology:
- Molecular cloning, protein production & engineering.
- Site-specific isotopic labeling (SSIL).
- Genetic code expansion (GCE) technology to incorporate non-canonical amino acids.
- Steady-state and time-resolved biophysical methods:
- Femtosecond-stimulated Raman spectroscopy (FSRS).
- 1D and 2D-infrared spectroscopy (2D-IR).
- Fluorescence spectroscopy (FRET, FCS and FA).
- Small-angle neutron (SANS) and X-ray scattering (SAXS).
- Computational methods:
- Density functional theory (DFT).
- Quantum mechanics / molecular mechanics (QM / MM).
- Molecular dynamics (MD).
Objectives
- Molecular basis of photocontrolled protein-protein and protein-DNA interactions.
- Development of GCE technology for the site-specific labeling of proteins.
- Analysis of photoinduced processes in biomolecules using advanced computational tools.
- Elucidation of vibrational energy transfer pathways in biomolecules.