About us
Cancer cells rewire their metabolism to satisfy the biosynthetic demands of rapid proliferation. As such cancer metabolism represents an attractive anti-cancer strategy. Historically, one of the first approaches to target cancer metabolism was anti-nucleotide therapy. More than 70 years old, this strategy blocks synthesis of nucleotides, the basic building blocks of DNA/RNA that are essential for proliferation. Despite its long history, anti-nucleotide therapy still suffers high rates of resistance and leads to toxicity in healthy tissues.
What are the reasons for resistance? While the answer to this question is not entirely known, at least part of the problem could be the exchange of nucleotides within tissues. Healthy tissues as well as malignant tumors consist of number of cell types with different metabolic needs and profiles. These cell types can metabolically communicate, e.g. one cell type can provide metabolites that are then taken up by another cell type in the same tissue. This crosstalk of metabolites enables collective survival in nutrient-poor environments. This may help tumor progression and metabolic crosstalk can thus limit the efficacy of metabolic interventions targeted at cancer cells.
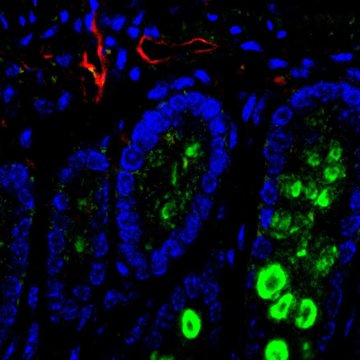
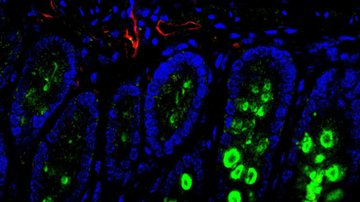
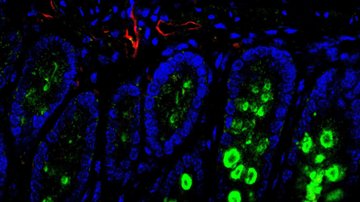
The cellular sources of nucleotides and their building blocks in tumors and in healthy tissues are not well understood. One of the reasons is that the traditional methods of metabolic research average populations and cannot differentiate individual cell types in their natural tissue environment.

The central goal of our research is to uncover how cells in tissues trade metabolites, and if healthy tissues and tumors differ in their metabolic ‘trading patterns’. Such knowledge and, especially the differences between healthy and tumor tissues, could be used to develop new therapeutic strategies that will overcome the resistance and toxicity.
To reach this goal we will combine genetic mouse models, metabolism and single cell technologies to map the metabolic crosstalk ‘one cell at a time’ in tumors as well as in healthy tissue. Specifically, our laboratory utilizes the state-of-the-art single cell approaches including single cell RNA-sequencing and spatial sequencing, combined with CRISPR screens and metabolic assays to investigate metabolic heterogeneity in tissues. Our experimental approach gives us a unique angle to study cancer and healthy tissue metabolism from a totally new perspective and with high resolution.