About us
We are a research team based at Charles University in Prague. We are evolutionary protistologists which means our interest is nothing less than origin and evolution of eukaryotic life. Our research focuses on two groups of organisms: Euglenida and Preaxostyla, both of which are members of Excavata supergroup.

We are particularly interested in the evolution of their unusual semi-autonomous organelles: highly reduced or even completely lost mitochondrion-like organelles of anaerobic Preaxostyla and secondary plastids of photosynthetic euglenids.
Both of these areas of research provide insight into organelle origin and evolution of their structure, molecular biology, transport, targeting, biogenesis, genome composition, molecular genetics mechanisms and biochemical pathways. We also study the phenomenon of lateral gene transfer, which plays an important role in some of these processes.
Preaxostyla
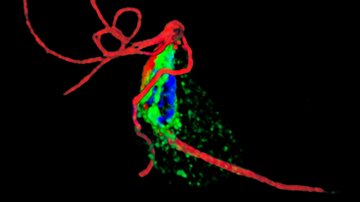
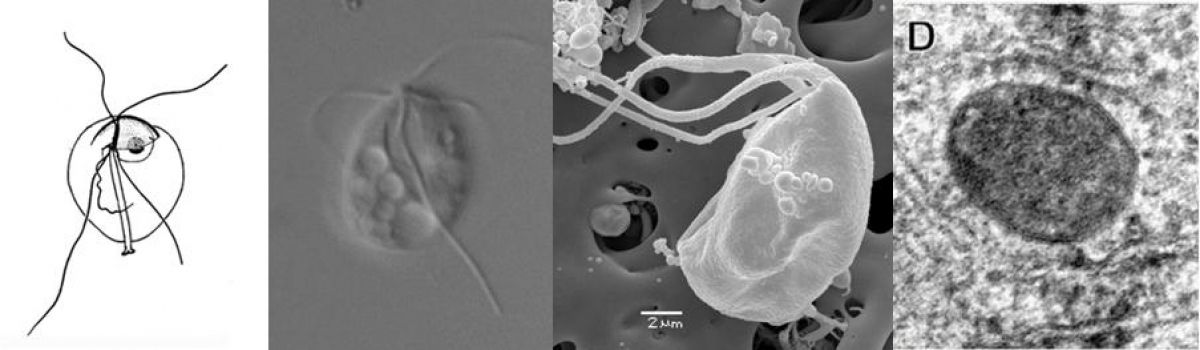
Preaxostyla, living exclusively in oxygen-depleted environments, are one of the least studied protist lineages. We believe these organisms can give us priceless insights into the reductive evolution of mitochondria. Paratrimastix pyriformis belongs to a basal assemblage of free-living Preaxostyla, formerly grouped under a single genus "Trimastix". We study the reduced mitochondrion of Paratrimastix in order to expose the physiological role of the organelle. Our second organisms of interest are oxymonads, which are all living inside the guts of various animals and are the largest known group of eukaryotes without any evidence of mitochondrion or related structures. Our investigation of the species Monocercomonoides exilis is focused on the intriguing possibility that this organism indeed completely lost the mitochondrion, a cellular structure that has been long thought to be essential for all eukaryotic organisms.
In our lab we focus on:
- transcriptomics and genomics of oxymonads and Paratrimastix
- cellular localization of potentially mitochondrial proteins
- metabolism connected with anaerobiosis and the putative loss of mitochondrion
- iron-sulfur clusters assembly pathways
- the symbiosis of oxymonads and their ecto- and endosymbiotic prokaryotes
- diversity of oxymonads
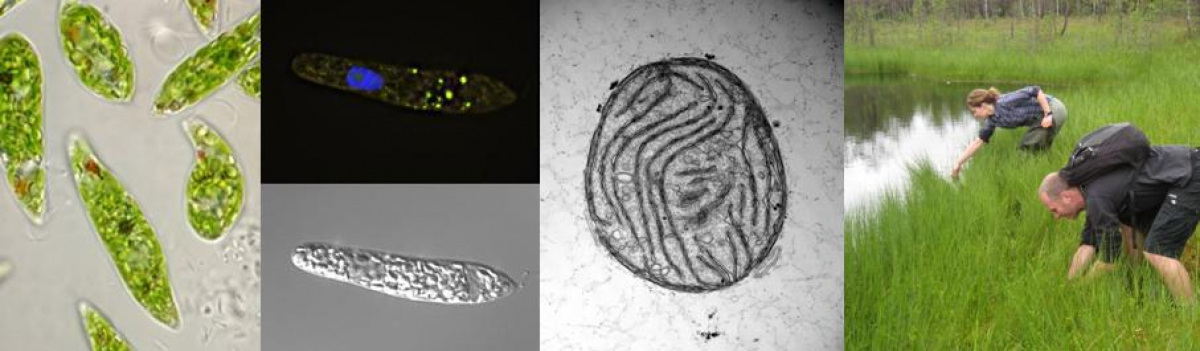
Euglenids
Euglenids are a group of mostly freshwater flagellates. Their mitochondria are unusual in both structure and molecular genetics. Many euglenids have firm but flexible pellicle and are capable of metaboly, a typical euglenoid movement. Euglenids are well known for their nutritional modes of diversity. The ancestral and most widespread mode of nutrition among euglenids is heterotrophy (bacteriovory, eukaryovory and primary osmotrophy). However, one monophyletic group, the euglenophytes, acquired a green secondary plastid and use photosynthesis as the main energy source. This plastid is derived from prasinophyte alga and has three envelope membranes. Light perceiving eyespot of unclear evolutionary origin is present in these organisms. The euglenophytes are still able to survive in dark by switching temporarily to heterotrophy; this feature enabled the origin of several secondarily osmotrophic species with non-photosynthetic colourless plastids. Rapaza viridis, recently discovered mixotrophic lineage requires both photosynthesis and eukaryotic prey for survival.
In our lab we focus on:
- transcriptomics and plastid genomics of euglenids and plastid-related algae
- lateral gene transfer that accompanied the origin of the plastid
- plastid proteomics and protein import in Euglena gracilis
- lipid analysis of euglenid chloroplast
- environmental sequencing in search for relatives of the plastid ancestor
- heterotrophic euglenids diversity
Currently, our research is conducted in these areas:
Eukaryotes with no mitochondria
- In 2016 we discovered that Monocercomonoides exilis cells lack mitochondria completely. It happens to be the very first eukaryotic organism, which provably lacks this organelle. Now we are trying to uncover the inner workings of such cells, and we are particularly interested in the synthesis of iron-sulfur clusters, which is a process closely related to mitochondria.
- We would like to characterize electron carrier proteins which take part in redox reactions in oxymonads and their relatives.
- We are also interested in the diversity of oxymonads and we would like to acquire a phylogenetic tree of oxymonads (as complete as possible). For this reason, we are collecting and sequencing oxymonads from their habitats.
- We want to try using the amitochondriate cells for "evolutionary" experiments to simulate the event of mitochondrial origin. For example, we are interested in what happens when we mix the cytosolic fraction of amitochondriate oxymonads with the isolated mitochondria of other organisms. Which proteins will be imported to the isolated mitochondria?
- Oxymonads might not be the only organisms without mitochondria. In collaboration with our colleague Ivan Čepička, we are investigating two other groups, which are potentially amitochondriate. Some of these are amoebas of the genus Pelomyxa and flagellates of the genus Retortamonas.
Euglenid plastids
- It is now clear, that the origin of the plastid of green euglenids is from secondary endosymbiosis with a green alga. It is also known that this alga was a relative to the genus Pyramimonas. Euglenid phylogeny suggests this event took place in marine environment. We do not know the age of such event, but it is evident that this endosymbiosis is younger than most of other endosymbiotic events.
- Our team wants to investigate the details of this event. We are interested in the evolution of plastid genomes, which is why we sequence these genomes in green algae and euglenids, which are phylogenetically closest to the endosymbiotic event. It's becoming apparent that the diversity of marine green and non-green euglenids is mapped only superficially, therefore we want to research it in depth. We hope for the discovery of new euglenid lineages.
- We are also interested in how the euglenid plastids function. In particular, we want to find out the origin of euglenid plastid membranes. These plastids have only three membranes (the ancestral number is four) and we want to know which membrane was lost in the evolution of euglenids. The mechanism of protein transport to euglenid plastids is also unclear. We selected several proteins, which could take part in the transport, however, their localisation has not been determined and we don't know their interacting partners.
- We would also like to learn how to transfect euglenas and suppress their gene expression by RNAi.
Protist cytoskeleton
- There is not much known about the protein composition of non-actin and non-tubulin cytoskeleton (intermediate and striated fibres) of protists. This knowledge might help us homologise morphological structures through divergent groups and help us answer some questions, such as how the last eukaryotic common ancestor (LECA) looked. Preliminary experiments show that we are able to prepare an enriched cytoskeleton fraction of oxymonad cytoskeleton and its subsequent proteomic analysis can tell us more about its composition.