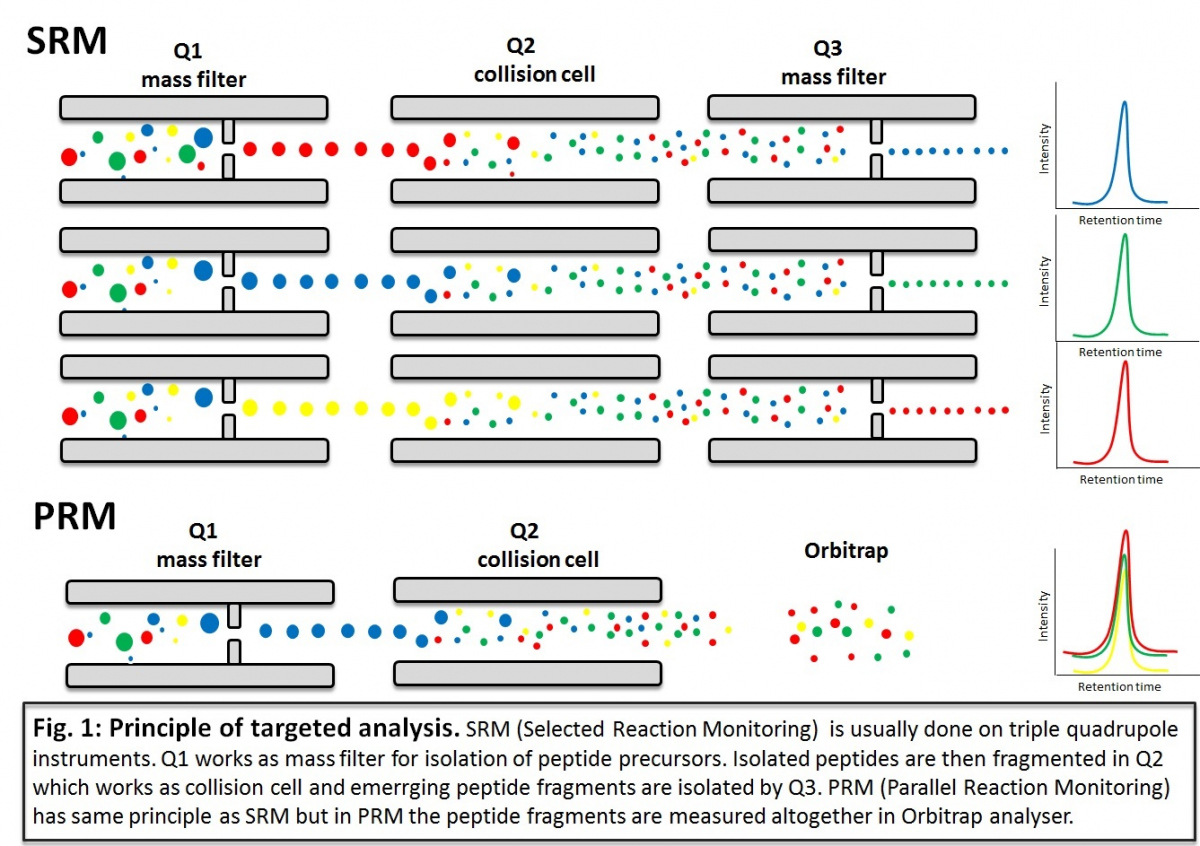
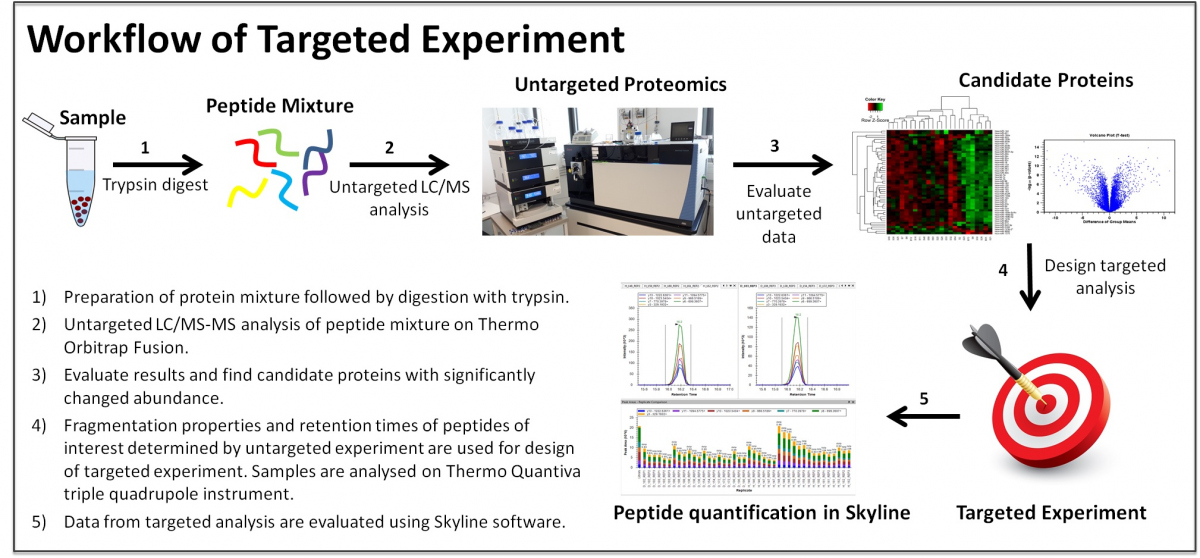
Ronald Malych, Pavel Stopka, Jan Mach, Eva Kotabová, Ondřej Prášil, Robert Sutak, Flow cytometry-based study of model marine microalgal consortia revealed an ecological advantage of siderophore utilization by the dinoflagellate Amphidinium carterae, Computational and Structural Biotechnology Journal, Volume 20, 2022, Pages 287-295, https://doi.org/10.1016/j.csbj.2021.12.023.
Vondráček O, Mikeš L, Talacko P, Leontovyč R, Bulantová J, Horák P. Differential proteomic analysis of laser-microdissected penetration glands of avian schistosome cercariae with a focus on proteins involved in host invasion. Int J Parasitol. 2022 Feb 23:S0020-7519(22)00017-0. doi: 10.1016/j.ijpara.2021.12.003. Epub ahead of print. PMID: 35218763.
Erban T, Klimov PB, Harant K, Talacko P, Nesvorna M, Hubert J. Label-free proteomic analysis reveals differentially expressed Wolbachia proteins in Tyrophagus putrescentiae: Mite allergens and markers reflecting population-related proteome differences. J Proteomics. 2021 Oct 30;249:104356. doi: 10.1016/j.jprot.2021.104356. Epub 2021 Aug 23. PMID: 34438106.
Vacurova E, Trnovska J, Svoboda P, Skop V, Novosadova V, Reguera DP, Petrezselyová S, Piavaux B, Endaya B, Spoutil F, Zudova D, Stursa J, Melcova M, Bielcikova Z, Werner L, Prochazka J, Sedlacek R, Huttl M, Hubackova SS, Haluzik M, Neuzil J. Mitochondrially targeted tamoxifen alleviates markers of obesity and type 2 diabetes mellitus in mice. Nat Commun. 2022 Apr 6;13(1):1866. doi: 10.1038/s41467-022-29486-z. PMID: 35387987.
Smutná T, Dohnálková A, Sutak R, Narayanasamy RK, Tachezy J, Hrdý I. A cytosolic ferredoxin-independent hydrogenase possibly mediates hydrogen uptake in Trichomonas vaginalis. Curr Biol. 2022 Jan 10;32(1):124-135.e5. doi: 10.1016/j.cub.2021.10.050. Epub 2021 Nov 10. PMID: 34762819.
Bláhová Z, Franěk R, Let M, Bláha M, Pšenička M, Mráz J. Partial fads2 Gene Knockout Diverts LC-PUFA Biosynthesis via an Alternative Δ8 Pathway with an Impact on the Reproduction of Female Zebrafish (Danio rerio). Genes (Basel). 2022 Apr 15;13(4):700. doi: 10.3390/genes13040700. PMID: 35456508; PMCID: PMC9032720.
Magalhaes-Novais S, Blecha J, Naraine R, Mikesova J, Abaffy P, Pecinova A, Milosevic M, Bohuslavova R, Prochazka J, Khan S, Novotna E, Sindelka R, Machan R, Dewerchin M, Vlcak E, Kalucka J, Stemberkova Hubackova S, Benda A, Goveia J, Mracek T, Barinka C, Carmeliet P, Neuzil J, Rohlenova K, Rohlena J. Mitochondrial respiration supports autophagy to provide stress resistance during quiescence. Autophagy. 2022 Oct;18(10):2409-2426. doi: 10.1080/15548627.2022.2038898. Epub 2022 Mar 8. PMID: 35258392; PMCID: PMC9542673.
Erban T, Sopko B, Bodrinova M, Talacko P, Chalupnikova J, Markovic M, Kamler M. Proteomic insight into the interaction of Paenibacillus larvae with honey bee larvae before capping collected from an American foulbrood outbreak: Pathogen proteins within the host, lysis signatures and interaction markers. Proteomics. 2022 Aug 10:e2200146. doi: 10.1002/pmic.202200146. Epub ahead of print. PMID: 35946602.
Volenikova, A., Nguyen, P., Davey, P. et al. Genome sequence and silkomics of the spindle ermine moth, Yponomeuta cagnagella, representing the early diverging lineage of the ditrysian Lepidoptera. Commun Biol 5, 1281 (2022). https://doi.org/10.1038/s42003-022-04240-9
Wu BC-h, Sauman I, Maaroufi HO, Zaloudikova A, Zurovcova M, Kludkiewicz B, Hradilova M and Zurovec M (2022) Characterization of silk genes in Ephestia kuehniella and Galleria mellonella revealed duplication of sericin genes and highly divergent sequences encoding fibroin heavy chains. Front. Mol. Biosci. 9:1023381. doi: 10.3389/fmolb.2022.1023381
Knizkova, D., Pribikova, M., Draberova, H. et al. CMTM4 is a subunit of the IL-17 receptor and mediates autoimmune pathology. Nat Immunol 23, 1644–1652 (2022). https://doi.org/10.1038/s41590-022-01325-9
Vdovin, A., Jelinek, T., Zihala, D. et al. The deubiquitinase OTUD1 regulates immunoglobulin production and proteasome inhibitor sensitivity in multiple myeloma. Nat Commun 13, 6820 (2022). https://doi.org/10.1038/s41467-022-34654-2
Pacakova L, Harant K, Volf P and Lestinova T (2022) Three types of Leishmania mexicana amastigotes: Proteome comparison by quantitative proteomic analysis. Front. Cell. Infect. Microbiol. 12:1022448. doi: 10.3389/fcimb.2022.1022448
Ujcikova, H.; Roubalova, L.; Lee, Y.S.; Slaninova, J.; Brejchova, J.; Svoboda, P. The Dose-Dependent Effects of Multifunctional Enkephalin Analogs on the Protein Composition of Rat Spleen Lymphocytes, Cortex, and Hippocampus; Comparison with Changes Induced by Morphine. Biomedicines 2022, 10, 1969. https://doi.org/10.3390/biomedicines10081969
Zimmann N, Rada P, Žárský V, Smutná T, Záhonová K, Dacks J, Harant K, Hrdý I, Tachezy J. Proteomic Analysis of Trichomonas vaginalis Phagolysosome, Lysosomal Targeting, and Unconventional Secretion of Cysteine Peptidases. Mol Cell Proteomics. 2022 Jan;21(1):100174. doi: 10.1016/j.mcpro.2021.100174. Epub 2021 Nov 8. PMID: 34763061; PMCID: PMC8717582.
Arbon D, Ženíšková K, Šubrtová K, Mach J, Štursa J, Machado M, Zahedifard F, Leštinová T, Hierro-Yap C, Neuzil J, Volf P, Ganter M, Zoltner M, Zíková A, Werner L, Sutak R. Repurposing of MitoTam: Novel Anti-Cancer Drug Candidate Exhibits Potent Activity against Major Protozoan and Fungal Pathogens. Antimicrob Agents Chemother. 2022 Aug 16;66(8):e0072722. doi: 10.1128/aac.00727-22. Epub 2022 Jul 20. PMID: 35856666; PMCID: PMC9380531.
Fry, M.Y., Najdrová, V., Maggiolo, A.O. et al. Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3. Nat Struct Mol Biol 29, 820–830 (2022). https://doi.org/10.1038/s41594-022-00798-4
Maria Grechnikova, Dominik Arbon, Kateřina Ženíšková, Ronald Malych, Jan Mach, Lucie Krejbichová, Aneta Šimáčková, Robert Sutak,
Elucidation of iron homeostasis in Acanthamoeba castellanii, International Journal for Parasitology, Volume 52, Issue 8, 2022, Pages 497-508, ISSN 0020-7519,
https://doi.org/10.1016/j.ijpara.2022.03.007.
Rada P, Hrdý I, Zdrha A, Narayanasamy RK, Smutná T, Horáčková J, Harant K, Beneš V, Ong S-C, Tsai C-Y, Luo H-W, Chiu C-H, Tang P and Tachezy J (2022) Double-Stranded RNA Viruses Are Released From Trichomonas vaginalis Inside Small Extracellular Vesicles and Modulate the Exosomal Cargo. Front. Microbiol. 13:893692. doi: 10.3389/fmicb.2022.893692
Petrusová, J.; Havalda, R.; Flachs, P.; Venit, T.; Darášová, A.; Hůlková, L.; Sztacho, M.; Hozák, P. Focal Adhesion Protein Vinculin Is Required for Proper Meiotic Progression during Mouse Spermatogenesis. Cells 2022, 11, 2013. https://doi.org/10.3390/cells11132013
Drastichova, Z.; Trubacova, R.; Novotny, J. β-Arrestin2 Is Critically Involved in the Differential Regulation of Phosphosignaling Pathways by Thyrotropin-Releasing Hormone and Taltirelin. Cells 2022, 11, 1473. https://doi.org/10.3390/cells11091473
Ronald Malych, Zoltán Füssy, Kateřina Ženíšková, Dominik Arbon, Vladimír Hampl, Ivan Hrdý, Robert Sutak, The response of Naegleria gruberi to oxidative stress, Metallomics, Volume 14, Issue 3, March 2022, mfac009, https://doi.org/10.1093/mtomcs/mfac009
Ženíšková K, Grechnikova M and Sutak R (2022) Copper Metabolism in Naegleria gruberi and Its Deadly Relative Naegleria fowleri. Front. Cell Dev. Biol. 10:853463. doi: 10.3389/fcell.2022.853463
Stemberkova-Hubackova S, Zobalova R, Dubisova M, Smigova J, Dvorakova S, Korinkova K, Ezrova Z, Endaya B, Blazkova K, Vlcak E, Brisudova P, Le DT, Juhas S, Rosel D, Daniela Kelemen C, Sovilj D, Vacurova E, Cajka T, Filimonenko V, Dong L, Andera L, Hozak P, Brabek J, Bielcikova Z, Stursa J, Werner L, Neuzil J. Simultaneous targeting of mitochondrial metabolism and immune checkpoints as a new strategy for renal cancer therapy. Clin Transl Med. 2022 Mar;12(3):e645. doi: 10.1002/ctm2.645. PMID: 35352502; PMCID: PMC8964933.
Erban, T., Shcherbachenko, E., Talacko, P. et al. Honey proteome of the bumblebee Bombus terrestris: similarities, differences, and exceptionality compared to honey bee honey as signatures of eusociality evolution. Apidologie 53, 16 (2022). https://doi.org/10.1007/s13592-022-00928-3
Jennifer J Tate, Jana Marsikova, Libuse Vachova, Zdena Palkova, Terrance G Cooper, Effects of abolishing Whi2 on the proteome and nitrogen catabolite repression-sensitive protein production, G3 Genes|Genomes|Genetics, Volume 12, Issue 3, March 2022, jkab432, https://doi.org/10.1093/g3journal/jkab432
Horáčková V, Voleman L, Hagen KD, Petrů M, Vinopalová M, Weisz F, Janowicz N, Marková L, Motyčková A, Najdrová V, Tůmová P, Dawson SC, Doležal P. Efficient CRISPR/Cas9-mediated gene disruption in the tetraploid protist Giardia intestinalis. Open Biol. 2022 Apr;12(4):210361. doi: 10.1098/rsob.210361. Epub 2022 Apr 27. PMID: 35472287; PMCID: PMC9042576.
Látrová K, Dolejšová T, Motlová L, Mikušová G, Bosák J, Snopková K, Šmajs D, Konopásek I, Fišer R. R-Type Fonticins Produced by Pragia fontium Form Large Pores with High Conductance. J Bacteriol. 2023 Jan 26;205(1):e0031522. doi: 10.1128/jb.00315-22. Epub 2022 Dec 21. PMID: 36541812; PMCID: PMC9879110.
Caspar KR, Stopka P, Issel D, Katschak KH, Zöllner T, Zupanc S, Žáček P, Begall S. Perioral secretions enable complex social signaling in African mole-rats (genus Fukomys). Sci Rep. 2022 Dec 26;12(1):22366. doi: 10.1038/s41598-022-26351-3. PMID: 36572727; PMCID: PMC9792591.
Zítek J, Füssy Z, Treitli SC, Peña-Diaz P, Vaitová Z, Zavadska D, Harant K, Hampl V. Reduced mitochondria provide an essential function for the cytosolic methionine cycle. Curr Biol. 2022 Dec 5;32(23):5057-5068.e5. doi: 10.1016/j.cub.2022.10.028. Epub 2022 Nov 7. PMID: 36347252; PMCID: PMC9746703.
Cejnar P, Smirnova TA, Kuckova S, Prochazka A, Zak I, Harant K, Zakharov S. Acute and chronic blood serum proteome changes in patients with methanol poisoning. Sci Rep. 2022 Dec 9;12(1):21379. doi: 10.1038/s41598-022-25492-9. PMID: 36494437; PMCID: PMC9734099.
Volenikova A, Nguyen P, Davey P, Sehadova H, Kludkiewicz B, Koutecky P, Walters JR, Roessingh P, Provaznikova I, Sery M, Zurovcova M, Hradilova M, Rouhova L, Zurovec M. Genome sequence and silkomics of the spindle ermine moth, Yponomeuta cagnagella, representing the early diverging lineage of the ditrysian Lepidoptera. Commun Biol. 2022 Nov 23;5(1):1281. doi: 10.1038/s42003-022-04240-9. PMID: 36418465; PMCID: PMC9684489.
Wu BC, Sauman I, Maaroufi HO, Zaloudikova A, Zurovcova M, Kludkiewicz B, Hradilova M, Zurovec M. Characterization of silk genes in Ephestia kuehniella and Galleria mellonella revealed duplication of sericin genes and highly divergent sequences encoding fibroin heavy chains. Front Mol Biosci. 2022 Nov 29;9:1023381. doi: 10.3389/fmolb.2022.1023381. PMID: 36523651; PMCID: PMC9745057.