About us
The Group of Reproductive biology focuses on characterization of processes and molecular factors involved in sperm maturation prior to fertilization (capacitation, acrosome reaction and seminal fluid composition), sperm-egg interaction, specification of anti-sperm antibodies, as well as developing tools for detection of male infertility.
The group is involved in monitoring of sperm quality in patients with testicular cancer and characterization of reproductive parameters modified by environmental factors with a negative effect on reproduction. This research area includes identification of epigenetic mechanisms participating in gametogenesis connected to gene expression. The goal of the group is to understand in detail localization and behaviour of selected protein prior and during the gamete fusion. The complex approach helps with a detection of pathologies and consequentially lead to prevention, diagnostics and selection of the most appropriate infertility treatment.

Our laboratory has extensive knowledge of assessing male reproductive parameters as markers of fertility disorders. We have been focusing on studying the molecular mechanisms of reproduction and the nature of specific sperm proteins that play a role in sperm maturation, sperm-egg interaction and early embryo development. The area of interest covers monitoring of sperm quality in patients with testicular cancer and diabetes mellitus and sperm antibodies characterization in infertile couples. Many of them are used in Centres of Assisted Reproduction and have been commercialized. Transgenerational epigenetic deregulation of microRNA expression induced by pollutants that influence the key role in germ cell differentiation without changing the DNA has been proven.
Recently, we have been studying in detail characterization of dynamic rearrangement of cytoskeletal proteins and their partner proteins (IZUMO1, CD46, CD9, CD81, CD151 and integrins) involved in sperm-egg fusion. In addition, the acrosin inhibitor expression and crucial ubiquitin proteasomal complex during sperm maturation has been addressed. Epigenetic aberrations (selected histone modifications and DNA methylation) in spermatozoa, testicular tissues and early embryos after exposure to environmental pollutants have been studied and new related project are in progress. All the ongoing research is supported by several prestigious grants. The outcome of this research is being translated into new diagnostic tools for identification of sperm parameters and selection of good and bad sperm to be used in Centres of Assisted Reproduction. This translational research is supported by grants of the Technological Agency of the Czech Republic and the Agency of Ministry of Health. Two European and two Czech patents were submitted.
We have productive collaboration based on the knowledge of the detailed assessment of reproductive parameters, monitoring of transgenerational epigenetic inheritance, characterization of epigenetic aberration and microRNA deregulations, all tailored for specific areas of interest (medical conditions, lifestyle, environmental factors). In addition, we utilize, in collaboration with commercial subjects, our knowledge of hybridoma technology for the production of specific monoclonal antibodies with high affinity, and twelve prototypes for the detection of relevant molecules have been developed. Moreover, we have valuable collaboration based on transgenic mouse lines, property of our laboratory, serving as indispensable tool for in vivo experiments, specifically transgenic C57BL/6Acr3-EGFP mice expressing green protein (EGFP) in sperm acrosome, transgenic C57BL/6Su9DsRed2 mice expressing red fluorescent protein (RFP) in somatic cell mitochondria.
ANNOTANTION OF MAJOR RESULTS:
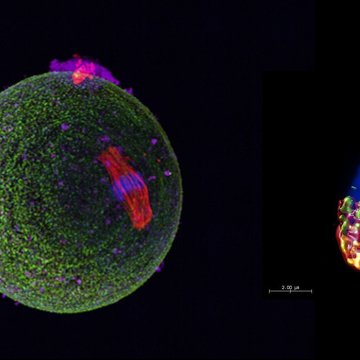
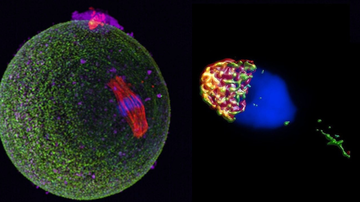
1. Characterization of CD46 and β1 integrin dynamics during sperm acrosome reaction.
Proteins CD46 and β1 integrin undergo synchronic dynamic relocation towards the sites of sperm-egg fusion during the acrosome reaction as shown by Super-resolution microscopy. Their interaction was confirmed by proximity ligation assay and their association with actin cytoskeleton was suggested. This protein relocation is suggested to be of importance for successful sperm-egg interaction, adhesion and subsequent gamete fusion and is being targeted as a tool for infertility diagnostics.
- Collaborating institutions: Department of Zoology, Faculty of Science, Charles University in Prague, 12844 Prague, Czech Republic.
- Contact person: Dr. Katerina Hortova, 604855871, katerina.hortova@ibt.cas.cz
- Publication: Frolikova M, Sebkova N, Ded L, Dvorakova-Hortova K. Characterization of CD46 and β1 integrin dynamics during sperm acrosome reaction. Sci Rep. 2016; 6:33714. doi: 10.1038/srep33714
2. CD9 and CD81 Interaction and their Structure Modelling in Sperm Prior to Fertilization.
The species-specific traits in CD9 and CD81 distribution during sperm maturation were compared between mice and humans. Confocal microscopy and colocalization assay was used to determine a mutual CD9/CD81 localization visualised in detail by super-resolution microscopy, and their interaction was address by co-immunoprecipitation. A mutual position of CD9/CD81 is shown in human spermatozoa in the acrosomal cap, however in mice, CD9 and CD81 occupy a distinct area. During the acrosome reaction in human sperm, only CD9 is relocated, compared to the relocation of both proteins in mice. The structural modelling of CD9 and CD81 homologous and possibly heterologous network formation was used to propose their lateral Cis as well as Trans interactions within the sperm membrane and during sperm-egg membrane fusion.
- Collaborating institutions: Department of Veterinary Sciences, Faculty of Agrobiology, Food and Natural Resources, University of Life Sciences Prague, Prague 6, 165 00, Czech Republic. Laboratory of Structural Bioinformatics of Proteins, Institute of Biotechnology CAS, v.v.i., BIOCEV, 252 50, Vestec, Czech Republic. Laboratory of Reproductive Physiology, Institute of Animal Biochemistry and Genetics Centre of Biosciences Slovak Academy of Sciences, 845 05 Bratislava, Slovakia. Department of Biochemistry and Department of Zoology, Faculty of Science, Charles University, Prague 2, 128 44, Czech Republic.
- Contact person: Dr. Katerina Hortova, 604855871, katerina.hortova@ibt.cas.cz
- Publication: Frolikova M, Manaskova-Postlerova P, Cerny J, Jankovicova J, Simonik O, Pohlova A, Secova P, Antalikova J, Dvorakova-Hortova K. CD9 and CD81 Interaction and their Structure Modelling in Sperm Prior to Fertilization. Int J Mol Sci. 2018 Apr 19;19(4). Pii: E1236. Doi: 10.3390/Ijms19041236.
3. Methylation analysis of histone H4K12ac-associated promoters in sperm of healthy donors and subfertile patients.
In men, histone-protamine exchange is incomplete, remaining histones are acetylated which represents an epigenetic mark transmitted from sperm and regulating early embryo gene expression. During mouse fertilization, paternal pronucleus exhibits a strong acetylation on H4K12, while maternal one shows its permanent increase until pronuclei fusion. Aberrant histone acetylation within developmentally important gene promoters in subfertile men but not DNA methylation, may reflect insufficient sperm chromatin compaction affecting the transfer of epigenetic marks to the egg.
- Collaborating institutions: Section Molecular Andrology, Biomedical Research Center Seltersberg, Justus Liebig University of Giessen, 35392 Giessen, Germany. Department of Zoology, Faculty of Science, Charles University in Prague, 12844 Prague, Czech Republic. Department of Urology, Pediatric Urology and Andrology, Justus Liebieg University of Giessen, 35392 Giessen, Germany. Fertility Center, 35578 Wetzlar, Germany. Fertility Center, 65189 Wiesbaden, Germany.
- Contact person: Mrs. Kateřina Hortová, 604855871, katerina.hortova@ibt.cas.cz
- Publication: Vieweg M, Dvorakova-Hortova K, Dudkova B, Waliszewski P, Otte M, Oels B, Hajimohammad A, Turley H, Schorsch M, Schuppe HC, Weidner W, Steger K, Paradowska-Dogan A.: Methylation analysis of histone H4K12ac-associated promoters in sperm of healthy donors and subfertile patients. Clin Epigenetics. 19;7(1):31. 2015 doi: 10.1186/s13148-015-0058-4.