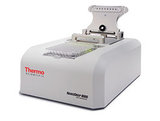
Droplet digital PCR with QX200 (Bio-Rad) for 8 – 96 samples
The ddPCR System provides absolute quantification of target DNA or RNA molecules for EvaGreen or probe-based digital PCR applications without a need for a standard curve. You can use 2 channels simultaneously (FAM, VIC).
We offer quantification with EvaGreen, probes and NGS libraries. All material is available at CF. Researchers can use instrument by themselves after training.
Droplet digital PCR principle:
- Droplet Digital PCR (ddPCR) is a method for performing digital PCR that is based on water-oil emulsion droplet technology. A sample is fractionated into 20,000 droplets.
- PCR amplification of the template molecules occurs in each individual droplet.
- Each droplet is analyzed or read in a flow cytometer to determine the fraction of PCR-positive droplets in the original sample. So, each positive droplet is calculated (1) and each negative droplet is calculated (0).These data are then analyzed using Poisson statistics to determine the target DNA template concentration in the original sample.

Aplications of ddPCR:
- Absolute quantification
- CNV (Copy number variation)
- Mutation analysis
- NGS validation and quantification of libraries
- Gene expression
- EvaGreen applications
Advantages of ddPCR:
- Absolute quantification — ddPCR technology provides an absolute count of target DNA copies per input sample without the need for running standard curves, making this technique ideal for measurements of target DNA, viral load analysis, and microbial quantification
- Unparalleled precision — the massive sample partitioning afforded by ddPCR enables the reliable measurement of small fold differences in target DNA sequence copy numbers among samples
- Increased signal-to-noise ratio — high-copy templates and background are diluted, effectively enriching template concentration in target-positive partitions, allowing for the sensitive detection of rare targets and enabling a ±10% precision in quantification
- Removal of PCR bias — error rates are reduced by removing the amplification efficiency reliance of qPCR, enabling the detection of small (1.2-fold) differences
- Simplified quantification — neither calibration standards nor a reference (the ΔΔCq method) is required for absolute quantification
- Superior partitioning — ddPCR technology yields 20,000 droplets per 20 µl sample, nearly two million partitioned PCR reactions in a 96-well plate, whereas chip-based digital PCR systems produce only hundreds or thousands of partitions. The greater number of partitions yields higher accuracy

ADDITIONAL RESOURCES:
http://www.bio-rad.com/en-cz/applications-technologies/droplet-digital-pcr-ddpcr-technology#1
https://www.youtube.com/watch?v=9TJy_uegmd4
https://www.thermofisher.com/cz/en/home/life-science/pcr/digital-pcr.html
Mikrofluidic digital PCR with BioMark (Fluidigm) for 12 or 48 samples
A sample in BioMark System is diluted and partitioned into hundreds or even millions of separate reaction chambers so that each contains one or no copies of the sequence of interest. By counting the number of ‘positive’ partitions (in which the sequence is detected) versus ‘negative’ partitions (in which it is not), scientists can determine exactly how many copies of a DNA molecule were in the original sample without need of a standard curve. What is more, you will obtain a real time PCR amplification curves and you can easily disqualify aberrant curves.
Microfluidic digital PCR principle:
- Sample is divided into individual compartments (1 sample into 770 compartments in a digital array)
- qPCR amplification of a specific amplicon using primers and probes (VIC and FAM channel)
- Positive and negative compartments, respectively, are calculated by software (1 or 0), software calculates with possibilities that there might be more copies of interest in one compartment (Poisson statistics). The target DNA template concentration in the original sample is determined.

Applications of dPCR:
- Absolute quantification
- CNV (Copy number variation)
- Mutation analysis
- EvaGreen applications
- Among other applications, researchers have used digital PCR to distinguish differential expression of alleles, to track which viruses infect individual bacterial cells, to quantify cancer genes in patient specimens and to detect fetal DNA in circulating blood

All material is available at CF
ADDITIONAL RESOURCES: https://www.fluidigm.com/search?query=digital+PCR&applications=digitalPCR